NC图片复现:多组学散点图(象限)
采用的是MASS包中的mvrnorm函数生成二元正态分布的数据,数据的列名分别为"mRNA_FC"和"RPF_FC"。
完整代码:
# 加载必要的库
library(MASS)
library(ggplot2)
library(cowplot)
# 生成数据
set.seed(123) # 设置随机种子以获得可复制的结果
covariance <- matrix(c(1,0.8,0.8,1),nrow=2,byrow=TRUE)
data <- mvrnorm(n=1000, mu=c(0,0), covariance)
data <- as.data.frame(data)
colnames(data) <- c("mRNA_FC", "RPF_FC")
# 分组处理
data$group <- ifelse(abs(data$mRNA_FC) > log10(2) & abs(data$RPF_FC) > log10(2),
"mRNA+RPF_both",
ifelse(abs(data$mRNA_FC) > log10(2),
"mRNA_only",
ifelse(abs(data$RPF_FC) > log10(2),
"RPF_only", NA)))
# 创建一个函数来绘制自定义的坐标轴
draw_axis_line <- function(length_x, length_y,
tick_step = NULL, lab_step = NULL) {
axis_x_begin <- -1 * length_x
axis_x_end <- length_x
axis_y_begin <- -1 * length_y
axis_y_end <- length_y
if (missing(tick_step))
tick_step <- 1
if (missing(lab_step))
lab_step <- 2
# 创建坐标轴刻度和标签的数据框
tick_x_frame <- data.frame(ticks = seq(axis_x_begin, axis_x_end, by = tick_step))
tick_y_frame <- data.frame(ticks = seq(axis_y_begin, axis_y_end, by = tick_step))
lab_x_frame <- subset(data.frame(lab = seq(axis_x_begin, axis_x_end, by = lab_step), zero = 0), lab != 0)
lab_y_frame <- subset(data.frame(lab = seq(axis_y_begin, axis_y_end, by = lab_step), zero = 0), lab != 0)
tick_x_length = 0.05
tick_y_length = 0.05
# 开始绘制图形
p <- ggplot() +
# 绘制坐标轴
geom_segment(aes(x = axis_x_begin, xend = axis_x_end, y = 0, yend = 0), size = 0.5) +
geom_segment(aes(x = 0, xend = 0, y = axis_y_begin, yend = axis_y_end), size = 0.5) +
# 添加刻度
geom_segment(data = tick_x_frame, aes(x = ticks, xend = ticks, y = 0, yend = -tick_x_length)) +
geom_segment(data = tick_y_frame, aes(x = 0, xend = -tick_y_length, y = ticks, yend = ticks)) +
# 添加刻度标签
geom_text(data = lab_x_frame, aes(x = lab, y = zero, label = lab), vjust = 1.5) +
geom_text(data = lab_y_frame, aes(x = zero, y = lab, label = lab), hjust = 1.5) +
theme_minimal() +
theme(panel.grid = element_blank(),
axis.text = element_blank())
return(p)
}
# 生成九宫格图
p <- draw_axis_line(4, 4)
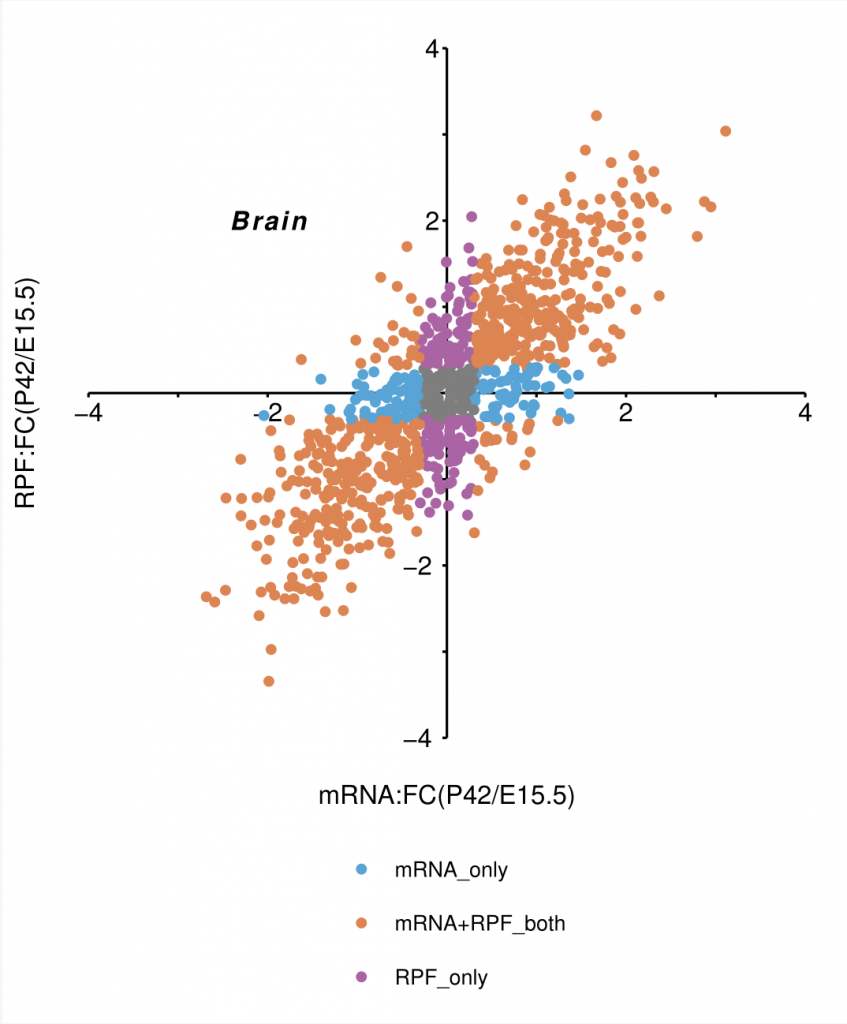
p1 <- p +
geom_point(data = data, aes(mRNA_FC, RPF_FC, color = group))+
scale_color_manual(values = c("mRNA+RPF_both" = "#dd8653", "mRNA_only" = "#59a5d7", "RPF_only" = "#aa65a4", "#878787")) +
labs(x = "mRNA:FC(P42/E15.5)", y = "RPF:FC(P42/E15.5)") +
theme(legend.position = "bottom") +
annotate("text", label = "bolditalic(Brain)", parse = TRUE, x = -2, y = 2, size = 4, colour = "black") +
guides(color = guide_legend(title = "", ncol = 1, byrow = TRUE))
# 保存图形为PDF
ggsave("plot.pdf", plot = p1, height = 6, width = 5)
# 拼接多个图形
p_list <- list(p1=p1, p2=p1, p3=p1)
p_all <- plot_grid(plotlist = p_list, ncol = 3)
# 保存拼接后的图形为PDF
ggsave("plot2.pdf", plot = p_all, height = 4, width = 9)
# 保存数据为CSV格式
write.csv(data, file = "data.csv", row.names = FALSE)
对于使用自己的数据,数据应该是一个csv文件,包含两列,列名分别为"mRNA_FC"和"RPF_FC"。其中每一行代表一个观测,"mRNA_FC"和"RPF_FC"是这个观测的两个特征值。
阅读剩余
版权声明:
作者:
链接:https://yunshangtulv.com.cn/?p=493
文章版权归作者所有,未经允许请勿转载。
THE END